Software
Stefania Marcotti, PhD
https://github.com/OakesLab/AFT-Alignment_by_Fourier_Transform
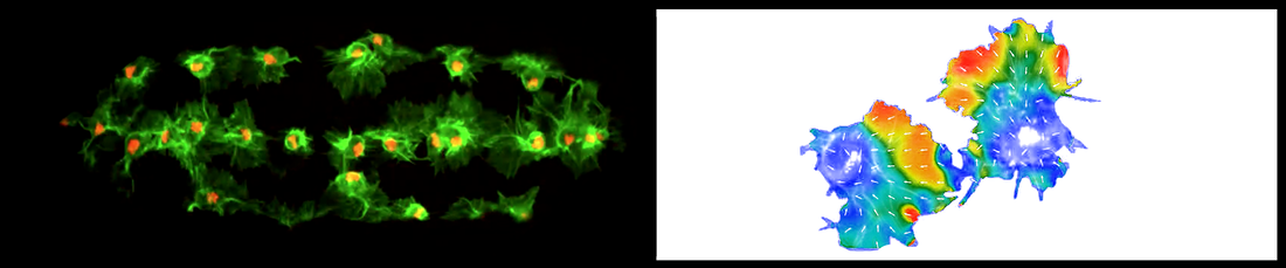
Quantifying alignment of fibrillar features
Open-source MATLAB and Python workflow to analyse alignment in bioimages. Highly flexible to different input types, it includes a tool for searching optimal analysis parameters and for evaluating the length scale of alignment. As always, we are here should you have any questions!
https://github.com/stemarcotti/protein_turnover_modelling
Modelling protein turnover
Have you read our recent publication on protein turnover (here!) and thought you would be curious to test the modelling on your data? We can help! We recently published a protocol paper in STAR Protocols (here!) to walk you through the analysis step by step. Have a look and get in touch should you have any questions!
https://github.com/stemarcotti/PIV
Particle Image Velocimetry (PIV) package developed in the Stramer Lab (King's College London, UK). Please see our publication on Nature Cell Biology (https://www.nature.com/articles/s41556-019-0411-5) for more details.
If you are interested in using this analysis tool on your data, please do get in touch with Brian or Stefania (see People and Contact pages for details)! We have extensive experience in adjusting the parameters for different kind of input movies to ensure that the analysis returns sensible results, and we will be more than happy to help.
https://github.com/stemarcotti/Fiji-macros
Miscellaneous of Fiji macros which might be helpful for batch analysis of images and movies (think saving all your open images, or run simple cell nuclei segmentation).
Do get in touch should you have any question or should anything not work as expected!
Quantifying alignment of fibrillar features
Open-source MATLAB and Python workflow to analyse alignment in bioimages. Highly flexible to different input types, it includes a tool for searching optimal analysis parameters and for evaluating the length scale of alignment. As always, we are here should you have any questions!
- Marcotti S*, Belo de Freitas D, Troughton LD, Kenny FN, Shaw TJ, Stramer B, Oakes PW* (2021). A workflow for rapid unbiased quantification of fibrillar feature alignment in biological images. Frontiers in Computer Science - Computer Vision. October 14, 2021 DOI: 10.3389/fcomp.2021.745831. * corresponding author
https://github.com/stemarcotti/protein_turnover_modelling
Modelling protein turnover
Have you read our recent publication on protein turnover (here!) and thought you would be curious to test the modelling on your data? We can help! We recently published a protocol paper in STAR Protocols (here!) to walk you through the analysis step by step. Have a look and get in touch should you have any questions!
- Marcotti S, Sánchez-Sánchez BJ, Serna-Morales E, Dragu A, Díaz-de-la-Loza MC, Matsubayashi Y, Stramer B. (2021). Protocol for intervention-free quantification of protein turnover rate by steady-state modeling. STAR Protocols. March 19, 2021 DOI: 10.1016/j.xpro.2021.100377
https://github.com/stemarcotti/PIV
Particle Image Velocimetry (PIV) package developed in the Stramer Lab (King's College London, UK). Please see our publication on Nature Cell Biology (https://www.nature.com/articles/s41556-019-0411-5) for more details.
If you are interested in using this analysis tool on your data, please do get in touch with Brian or Stefania (see People and Contact pages for details)! We have extensive experience in adjusting the parameters for different kind of input movies to ensure that the analysis returns sensible results, and we will be more than happy to help.
https://github.com/stemarcotti/Fiji-macros
Miscellaneous of Fiji macros which might be helpful for batch analysis of images and movies (think saving all your open images, or run simple cell nuclei segmentation).
Do get in touch should you have any question or should anything not work as expected!
Methods
Working with the Drosophila embryo
In our lab, many projects use Drosophila embryogenesis to study the role and regulation of cell migration in animal physiology, and mechanisms controlling extracellular matrix formation. To look at the fly embryo in real time, we have developed a variety of live imaging techniques, using a variety of microscopy systems. Here, there are some links to our more frequently used protocols, from embryo collection to imaging.